End-to-End Guide: Fine-Tuning Embeddings for Domain-Specific RAG
Overview
Retrieval-Augmented Generation (RAG) has changed the way we build intelligent applications. Instead of relying only on what a language model already knows, RAG lets it pull answers from private or highly specific documents. The engine behind this process is the embedding model—the quiet workhorse that turns text into numerical vectors so the system can quickly find what matters.
General-purpose embedding models, like BAAI/bge-base-en-v1.5, are strong performers out of the box. But drop them into specialized fields and they start to miss the mark. Legal contracts, scientific research, or FDA regulatory filings all come with their own vocabulary and subtle distinctions that a generic model may not fully understand.
The solution? Fine-tuning.
By training an embedding model on domain-specific data, we help it learn the language of our documents. The payoff is simple: sharper retrieval, more relevant answers, and stronger RAG results.
Most resources you will find on the internet about fine-tuning use ready-made datasets. That works for demos or learning, but in real enterprise settings you rarely get that convenience. Data usually comes in PDFs, Word files, or messy internal knowledge bases. In this post we take the practical route: building a clean dataset from scratch using raw FDA documents, then fine-tuning on it.
In this blog, we will cover:
Data Preparation: Chunking complex documents that models can actually work with.
Synthetic Data Generation: Using
gpt-4o-minito create our training dataset.Baseline Evaluation: Figuring out where we stand before fine-tuning, so we can show the improvements clearly.
The Fine-Tuning Process: Using the
sentence-transformerslibrary and Multiple Negatives Ranking Loss (MNRL).Final Evaluation & Comparison: Quantifying the improvements and visualizing the results..
Preparing Domain-Specific Data
The quality of data makes or breaks an AI model. For fine-tuning, that meant turning raw FDA guidance documents (scraped from FDA.gov and converted from PDF to Markdown) into clean, well-structured chunks of text.
Chunking isn't just about splitting text; it's about creating meaningful units of information. A poorly chunked document, for example, splitting a sentence in half, can confuse the embedding model. Our strategy is multi-layered:
Preprocessing: We use regular expressions to strip out Markdown syntax (like ##, **, and links). We want the model to learn the meaning of the text, not the formatting.
Recursive Splitting: We use recursive character text splitter to intelligently split the text, prioritizing paragraph breaks over sentence breaks to keep related ideas together.
Sentence-Level Refinement: For any chunks that are still too large, we split them into sentences and group them back together, ensuring we never exceed our token limit or create awkward mid-sentence breaks.
Metadata: We attach crucial metadata to each chunk, like the source file and chunk number. That way, we can always trace back to the original and even provide citations.
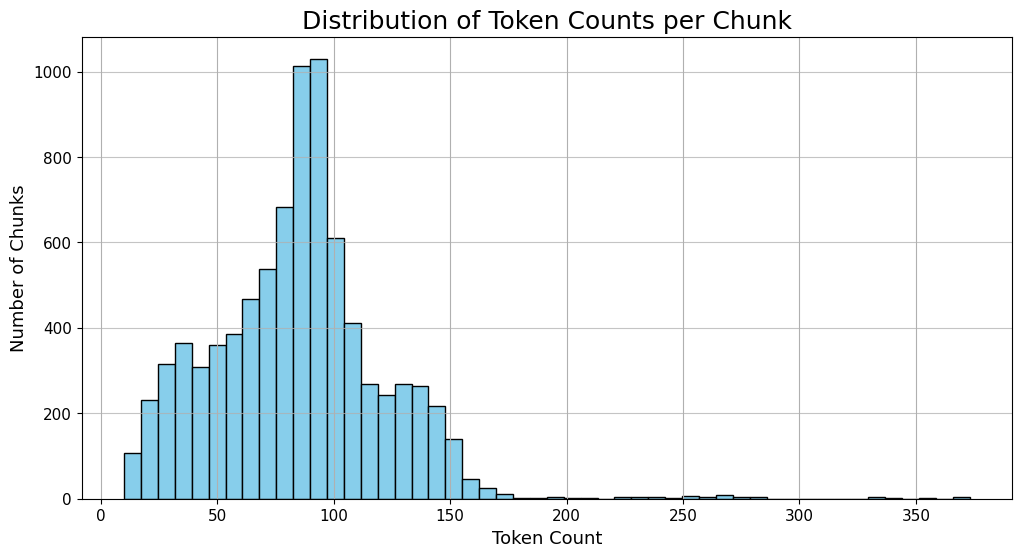
Once chunking was done, we checked the size distribution to make sure everything lined up well with the model’s sweet spot.
The graph highlights the payoff of our chunking approach. Most chunks fall in the 80–95 token range—big enough to carry meaningful context, yet small enough to stay precise. That sweet spot is exactly what makes a RAG system effective.
Synthetic Data Generation
To fine-tune our model, we need question-answer pairs. Since we don't have a pre-existing dataset of researcher questions about FDA documents, we'll create one. This is where synthetic data generation comes in.
Our goal is to generate high-quality questions that are specific to a single chunk and answerable only by its content. For this, we turn to gpt-4o-mini and the instructor library.
The instructor library allows us to get structured, Pydantic-validated output from OpenAI's API. Instead of parsing messy strings, we can define the exact Python object we want back.
Here’s the Pydantic model we use to define our desired output:
from pydantic import BaseModel, Field
class Question(BaseModel):
"""Schema for a single question generated by the AI for a chunk."""
question: str = Field(..., description="A highly specific question that is answerable only by the given text chunk.")
Next, we build a system prompt to guide the model. The prompt tells it to act as a medical device regulation expert, generate one clear question from each chunk, and return “None” if the chunk isn’t useful, like contact details or a table of contents. This safeguard keeps the dataset focused and raises its overall quality.
Here's a snippet of the system prompt:
SYSTEM_PROMPT = """
You are an expert in medical device regulation and clinical trials. Your task is to act as a researcher or device developer working through an FDA guidance document.
From each text chunk provided:
1. **Detect Conceptual Guidance:** First verify the chunk contains at least one conceptual or “actionable” statement about device-study design, data analysis, or regulatory expectations (e.g. sentences describing purposes, justifications, recommendations, requirements, or rationale).
2. **Generate One Question or None:**
- If step 1 succeeds, craft exactly one clear, answerable question that a researcher would ask to deepen understanding of that concept or recommendation.
- If step 1 fails (the chunk is only metadata, contact details, legal boilerplate, unrelated filler, or lacks any conceptual guidance), output exactly:
None
3. **Researcher Persona & Depth:** The question must sound like a human researcher’s curiosity—focusing on “how” or “why,” not mere fact-recall—and it must be fully answerable *using only* the provided text.
4. **Avoid Trivia:** Don’t ask about dates, document numbers, or contacts unless those details *are* the one and only conceptual point in the chunk.
5. **Be Specific:** Anchor the question in the chunk’s wording and scope.
**Weak example (should yield None):**
> Chunk only contains “Issued on June 20, 2016. Contact …”
**Good example:**
> Chunk states “The guidance recommends stratifying clinical outcomes by age and race to detect subgroup effects.”
> → “Why does the guidance recommend stratifying clinical outcomes by age and race when evaluating medical device performance?”
"""
By running our 8,361 chunks through this process, we generated over 5,700 high-quality question-chunk pairs, which we then split into training (90%) and evaluation (10%) sets.
The Baseline Model
You can't know if you've improved if you don't know where you started. Before fine-tuning, we must establish a baseline by measuring the performance of the off-the-shelf BAAI/bge-base-en-v1.5 model on our evaluation data.
The process is straightforward:
Embed our entire corpus of 8,361 document chunks using the base model.
Store these embeddings in a ChromaDB vector database.
For each question in our evaluation set, embed it and search the database for the most similar chunks.
Measure the performance using standard information retrieval metrics.
Key Metrics Explained:
Recall@k: Did we find the correct document chunk within the top k results? A Recall@5 of 0.90 means that 90% of the time, the right answer was in the top 5 retrieved documents.
Mean Reciprocal Rank (MRR@k): How high up in the rankings was the correct document? This metric heavily rewards models that place the correct answer at the very top.
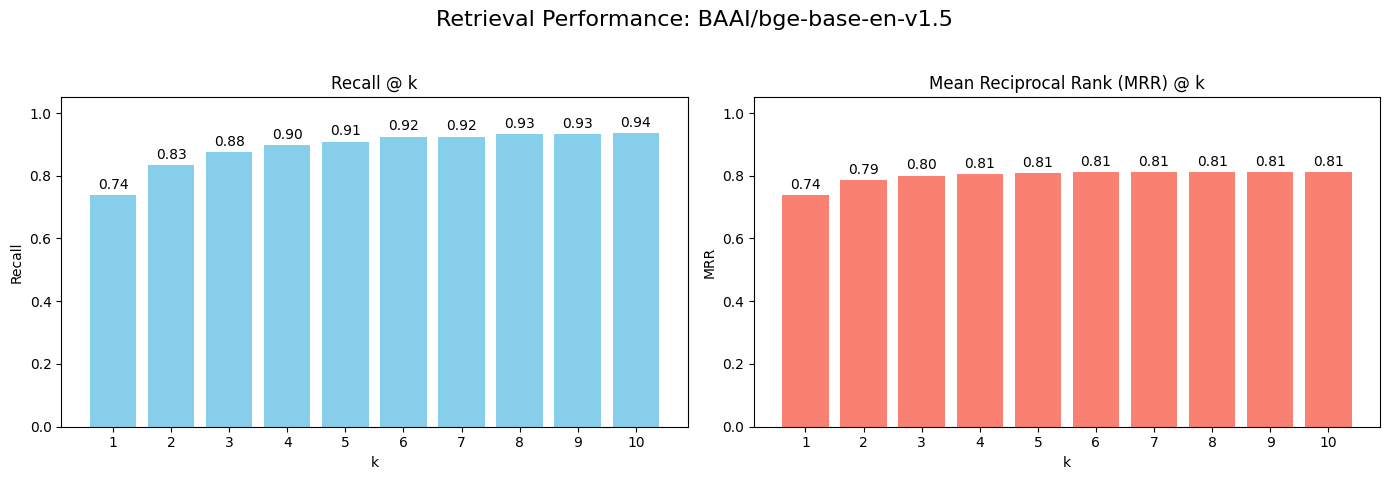
Here are the results for our baseline model:
These are strong starting numbers, but there's room for improvement. A Recall@1 of ~74% means that for one in four questions, the single best answer isn't the first result. We can do better.
Fine-Tuning with MNRL
Now for the exciting part. We'll use our synthetic training data to adapt the BAAI/bge-base-en-v1.5 model to the specific nuances of FDA documents.
Our strategy is Multiple Negatives Ranking Loss (MNRL). This training technique is perfect for our (question, chunk) data format. In simple terms, for each training batch, the model learns to:
Maximize the similarity between a question and its correct "positive" chunk.
Minimize the similarity between that same question and all other "negative" chunks in the batch.
By repeating this thousands of time, the model learns to create a vector space where questions are located extremely close to their corresponding answers.
The sentence-transformers library makes this complex process surprisingly simple. The core of our training script is the model.fit() call:
model.fit(
train_objectives=[(train_loader, train_loss)],
evaluator=evaluator,
epochs=EPOCHS,
warmup_steps=warmup_steps,
output_path=str(FT_MNRL_PATH),
save_best_model=True,
use_amp=True,
optimizer_params={'lr': LEARNING_RATE}
)
We ran it for 3 epochs with learning rate of 2e-5.
Was It Worth It?
This is the moment of truth. We repeat the exact same evaluation process as before, but this time using our newly fine-tuned model to generate the embeddings.
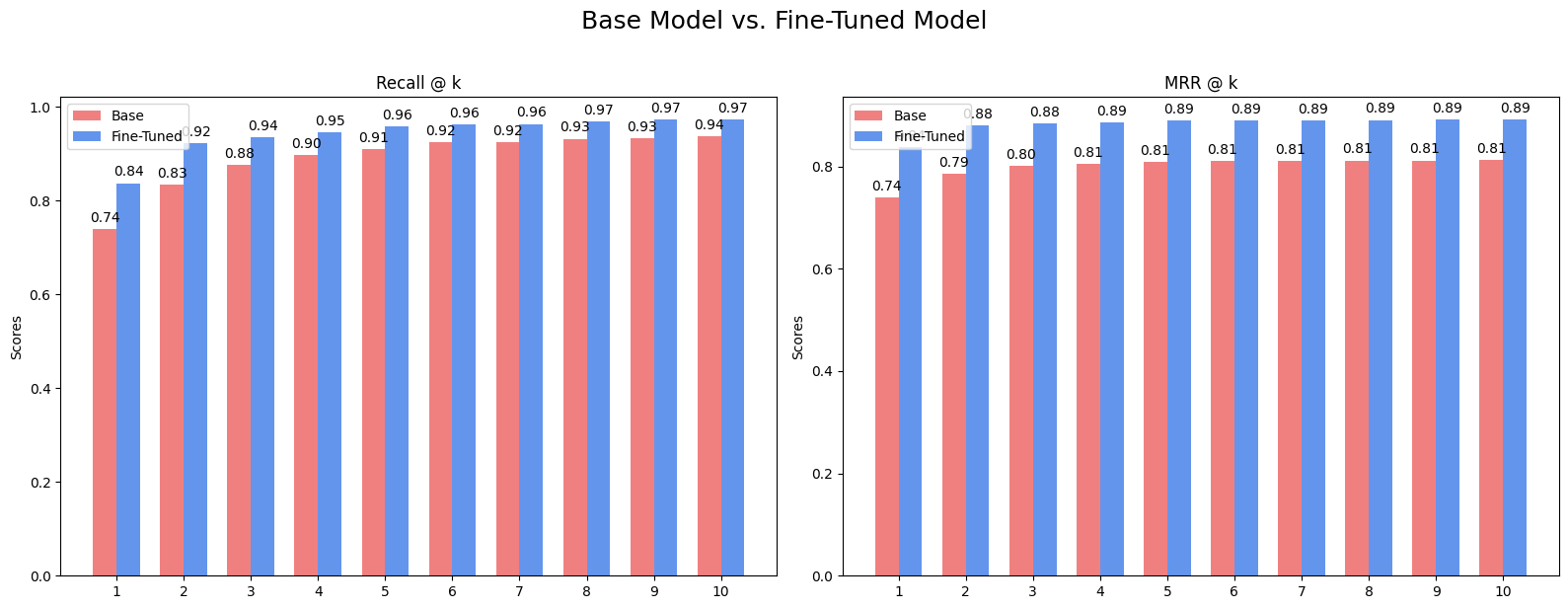
The improvements are significant across the board. Recall@1 jumped from 74% to nearly 84%. This means our RAG system is now our RAG system is now about 10% more likely to put the best document in the very first spot. The Mean Reciprocal Rank (MRR) also saw a substantial lift, indicating that even when the correct answer isn't #1, it's ranked much higher than before.
This visual comparison makes the impact even clearer:
This bar chart visually contrasts the performance, showing the fine-tuned model (in blue) consistently outperforming the base model (in Salmon) for both Recall@k and MRR@k across all values of k.
Conclusion
Fine-tuning is not black magic; it's a systematic process that can yield dramatic performance gains for domain-specific RAG systems. By carefully preparing our data, leveraging powerful LLMs to generate a synthetic dataset, and using a well-established training technique like MNRL, we successfully taught a general-purpose model the specific language of FDA regulatory documents.
The techniques outlined here are not limited to FDA documents. They can be applied to any specialized domain - legal texts, financial reports, engineering manuals, or internal company wikis - to build smarter, more accurate, and more reliable RAG applications.
For the complete, runnable code, you can find the full Python notebook here.